Almost all cells of an organism have the same DNA. When a cell divides, this DNA is passed on to the new cells. Sometimes, a mutation to the DNA occurs. When this happens, the cell function changes. This is called a somatic mutation. In what sense the function of a cell changes when its DNA changes depends on the changes, where in the DNA it occurred etc. In most circumstances there will be little noticeable change or if there is a large change, the cell may not function anymore at all. Cells that die are shed through the bloodstream.
What is ctDNA?
Circulating tumor DNA (ctDNA) is a subset of cell-free DNA (cfDNA) — short DNA fragments that float freely in the blood. While cfDNA is released by many cells in the body (especially when cells die and are shed through the blood stream), ctDNA originates specifically from tumor cells and often carries genetic mutations that drive cancer.
By sequencing ctDNA and comparing it to the normal genome, it is sometimes possible to detect somatic mutations in important cancer-related genes (oncogenes and tumor suppressors), helping to diagnose or monitor cancer without a traditional biopsy. Even though it could be better to take a sample from a tumor.
Somatic mutations may not be very prevalent in the sample. Especially if there are not many cells in the body (yet) that have the mutation and you are measuring a blood sample. If you are interested in any somatic mutations in such a sample, the higher the coverage of the sequencing the better. The chance of measuring a mutation that is not very prevalent in the sample is higher if the number of reads (the coverage) is higher. A coverage of 30X could still not sequence any somatic mutations in that case and a higher coverage is recommended.
For an example of its clinical potential in a practical situation, please see this recent publication:
🔗 Circulating Tumor DNA Analysis for Solid Tumors (2025)
ISLB (International Society for Liquid Biopsy) — Leading research and clinical guidelines
Tumor Size and ctDNA Levels
The amount of ctDNA in the blood generally reflects the size and activity of the tumor. In early-stage cancers, the tumor burden is small, meaning fewer cancer cells are present — and as a result, ctDNA levels are often very low. This can make detection more challenging and may require highly sensitive sequencing and analysis techniques.
In contrast, advanced or fast-growing tumors tend to shed more ctDNA, making mutations easier to detect in a blood sample.
This dynamic makes liquid biopsy especially useful for:
- Monitoring treatment response (as ctDNA levels rise or fall)
- Identifying recurrence earlier than imaging alone
What are Onco-Mutations?
Onco-mutations are specific changes in DNA that could contribute to the development and progression of cancer. These mutations mostly occur in genes that regulate cell growth, division, and repair. When such genes are altered, the normal control mechanisms of the cell cycle can break down, leading to uncontrolled cell proliferation—a hallmark of cancer.
Types of Genetic Mutations in Cancer:
- Oncogenes – When mutated or abnormally expressed, these genes can drive the transformation of a normal cell into a cancerous one.
- Tumor Suppressor Genes – These normally act as brakes on cell growth. Mutations can disable this function.
- DNA Repair Genes – These help fix errors in DNA. Mutations in these genes can lead to accumulation of additional mutations.
Why They Matter
Understanding these mutations can help in diagnosing cancer, predicting its behavior, and choosing the most effective treatment. Many targeted therapies are designed to counteract specific mutations. We have listed some approved, some experimental therapies here.
What this tool searches
This tool searches in the datafile you provide for large differences with a reference (i.e. healthy) DNA sequence. If found, it will list the exact name and location of the mutation. This can then be used for further research into the specific conditions of the patient.
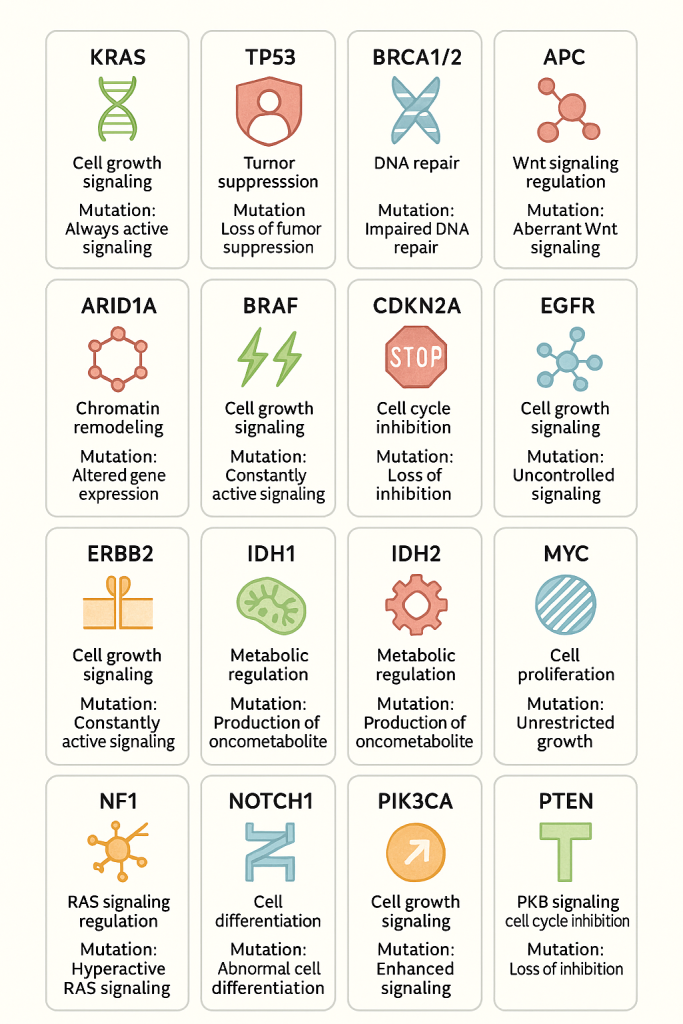
Oncogene & Tumor Suppressor General Overview
1. KRAS
- Function: Encodes a GTPase that regulates cell proliferation via RAS/MAPK signaling.
- Prevalence: Mutations are common in pancreatic (up to 90%), colorectal (30–40%), and lung cancers (~25%).
- Mechanism: Mutated KRAS is constitutively active, driving continuous cell division without external growth signals.
2. TP53
- Function: Tumor suppressor; regulates DNA repair, apoptosis, and cell cycle arrest.
- Prevalence: Mutated in over 50% of human cancers.
- Mechanism: Loss of function leads to unchecked cell division and resistance to apoptosis.
3. BRCA1/2
- Function: Involved in homologous recombination repair of DNA double-strand breaks.
- Prevalence: Germline mutations in 1 in 400–800 people; high in hereditary breast and ovarian cancers.
- Mechanism: Impaired DNA repair leads to genomic instability and tumor development.
4. APC
- Function: Regulates Wnt signaling and controls cell adhesion and migration.
- Prevalence: Mutated in ~80% of colorectal cancers.
- Mechanism: Loss leads to Wnt pathway activation and unregulated cell proliferation.
5. ARID1A
- Function: Chromatin remodeling for gene expression regulation.
- Prevalence: Mutated in ~30–50% of ovarian clear cell and endometrial cancers.
- Mechanism: Alters chromatin structure, affecting gene transcription and cell cycle control.
6. BRAF
- Function: Serine/threonine kinase in the MAPK signaling pathway.
- Prevalence: V600E mutation in ~50% of melanomas, also in colorectal and thyroid cancers.
- Mechanism: Activates MAPK signaling independent of upstream signals, promoting proliferation.
7. CDKN2A
- Function: Produces p16, an inhibitor of cyclin-dependent kinases; controls G1/S checkpoint.
- Prevalence: Frequently inactivated in melanoma, pancreatic, and glioblastomas.
- Mechanism: Loss removes a key cell cycle checkpoint, allowing uncontrolled cell division.
8. EGFR
- Function: Receptor tyrosine kinase that triggers growth and survival pathways.
- Prevalence: Mutations common in non-small cell lung cancer (NSCLC) and glioblastomas.
- Mechanism: Constitutively active signaling supports growth and evades apoptosis.
9. ERBB2 (HER2)
- Function: Receptor tyrosine kinase involved in cell proliferation.
- Prevalence: Amplified in ~20% of breast cancers, also in gastric and esophageal cancers.
- Mechanism: Overexpression leads to excessive growth signaling.
10. IDH1/IDH2
- Function: Enzymes in citric acid cycle; produce α-ketoglutarate.
- Prevalence: Mutated in gliomas, AML, and cholangiocarcinomas.
- Mechanism: Mutants produce 2-hydroxyglutarate, an oncometabolite that inhibits differentiation.
11. MYC
- Function: Transcription factor that promotes cell growth and division.
- Prevalence: Amplified in many cancers (e.g., Burkitt lymphoma, breast cancer).
- Mechanism: Drives rapid proliferation and metabolic reprogramming.
12. NF1
- Function: Tumor suppressor; negative regulator of RAS signaling.
- Prevalence: Mutated in neurofibromatosis type 1, gliomas, and MPNST.
- Mechanism: Loss enhances RAS signaling, promoting cell growth.
13. NOTCH1
- Function: Regulates cell fate decisions and differentiation.
- Prevalence: Mutated in T-cell ALL, head and neck cancers.
- Mechanism: Gain- or loss-of-function mutations alter differentiation, often leading to uncontrolled proliferation.
14. PIK3CA
- Function: Catalytic subunit of PI3K, activates AKT/mTOR pathway.
- Prevalence: Mutations in ~30% of breast and endometrial cancers.
- Mechanism: Enhances survival and growth signals.
15. PTEN
- Function: Tumor suppressor; antagonizes PI3K/AKT signaling.
- Prevalence: Mutated in glioblastoma, prostate, and endometrial cancers.
- Mechanism: Loss of PTEN increases AKT activity, promoting survival and proliferation.
16. RB1
- Function: Controls G1/S transition via E2F inhibition.
- Prevalence: Inactivated in retinoblastoma, osteosarcoma, small-cell lung cancer.
- Mechanism: Without RB1, cells bypass G1/S checkpoint, leading to uncontrolled division.
